Simulated single cell expression data with dropout¶
This example uses TACCO to annotate simulated single cell data, simulated using a modified variant (Moriel) of scsim (Kotliar).
(Moriel): Moriel, N. Extension of scsim single-cell RNA-sequencing data simulations. github.com/nitzanlab/scsim-ext (2023)
(Kotliar): Kotliar, D. scsim: simulate single-cell RNA-SEQ data using the Splatter statistical framework but implemented in python. github.com/dylkot/scsim (2021)
[1]:
import os
import sys
import matplotlib
import pandas as pd
import numpy as np
import scipy.sparse
import scanpy as sc
import tacco as tc
from scsim import scsim
# The notebook expects to be executed either in the workflow directory or in the repository root folder...
sys.path.insert(1, os.path.abspath('workflow' if os.path.exists('workflow/common_code.py') else '..'))
import common_code
[2]:
env_path = common_code.find_path('results/env_links')
Simulate single cell data¶
Simulate data with various dropout levels with a known ground truth
[3]:
ngenes = 25000
descale = 1.0
ndoublets = 100
K = 13
nproggenes = 1000
proggroups = [1,2,3,4]
progcellfrac = .35
ncells = 1500
deprob = .025
seed = 111
deloc = 2.0
# simulating true counts (in simulator.counts)
simulator = scsim(ngenes=ngenes, ncells=ncells, ngroups=K, libloc=7.64, libscale=0.78,
mean_rate=7.68,mean_shape=0.34, expoutprob=0.00286,
expoutloc=6.15, expoutscale=0.49,
diffexpprob=deprob, diffexpdownprob=0., diffexploc=deloc, diffexpscale=descale,
bcv_dispersion=0.448, bcv_dof=22.087, ndoublets=ndoublets,
nproggenes=nproggenes, progdownprob=0., progdeloc=deloc,
progdescale=descale, progcellfrac=progcellfrac, proggoups=proggroups,
minprogusage=.1, maxprogusage=.7, seed=seed)
simulator.simulate()
Simulating cells
Simulating gene params
Simulating program
Simulating DE
Simulating cell-gene means
- Getting mean for activity program carrying cells
- Getting mean for non activity program carrying cells
- Normalizing by cell libsize
Simulating doublets
Adjusting means
Simulating counts with scsim
[4]:
reference = sc.AnnData(scipy.sparse.csr_matrix(simulator.counts), obs=simulator.cellparams, var=simulator.geneparams)
reference.obs['group'] = reference.obs['group'].astype('category')
# fitting number of zeros vs log mean curve
dropshape, dropmidpoint = simulator.fit_dropout()
dropout_midpoints = [1.0,-1.0,-3.0,-5.0,-7.0]
# to add technical dropout noise, we can decrease the sigmoid's dropmidpoint to increase the dropout (in simulator.countswdrop)
sdatas = {}
for dropout_midpoint in dropout_midpoints:
simulator.dropshape = dropshape
simulator.dropmidpoint = dropout_midpoint
simulator.simulate_dropouts()
sdata = sc.AnnData(scipy.sparse.csr_matrix(simulator.countswdrop), obs=simulator.cellparams, var=simulator.geneparams)
sdata.obs['group'] = sdata.obs['group'].astype('category')
sdatas[dropout_midpoint] = sdata
Plotting options¶
[5]:
highres = False
default_dpi = 100.0
if highres:
matplotlib.rcParams['figure.dpi'] = 648.0
hr_ext = '_hd'
else:
matplotlib.rcParams['figure.dpi'] = default_dpi
hr_ext = ''
axsize = np.array([3,3])*1.0
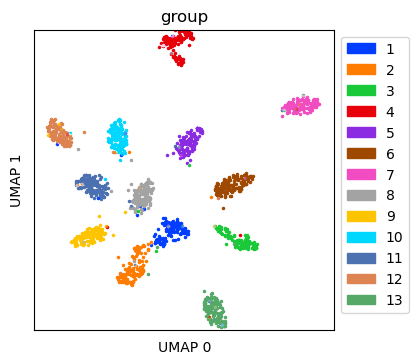
Visualize simulated data without dropout¶
Create UMAP for the simulated data without dropout
[6]:
ref_umap = tc.utils.umap_single_cell_data(reference)
fig = tc.pl.scatter(ref_umap, keys='group', position_key='X_umap', joint=True, point_size=5, axsize=axsize, noticks=True,
axes_labels=['UMAP 0','UMAP 1']);
SCumap...SCprep...time 1.4392147064208984
time 26.896665811538696
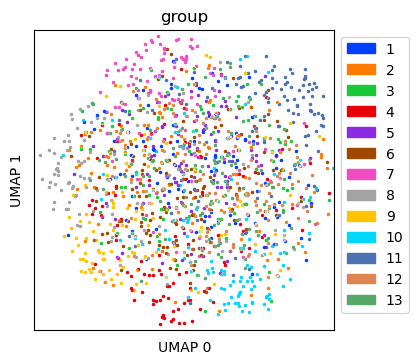
Visualize simulated data with dropout¶
Create UMAP for the simulated data with dropout
[7]:
tdata = sdatas[-1.0]
tdata_umap = tc.utils.umap_single_cell_data(tdata)
fig = tc.pl.scatter(tdata_umap, keys='group', position_key='X_umap', joint=True, point_size=5, axsize=axsize, noticks=True,
axes_labels=['UMAP 0','UMAP 1']);
SCumap...SCprep...time 0.5789980888366699
time 8.304261207580566
Annotate the data with dropout with cell types¶
Annotation is done on cell type level with multi_center=10 to capture variation within a cell type.
[8]:
tc.tl.annotate(tdata, reference, 'group', result_key='TACCO', multi_center=10, assume_valid_counts=True)
Starting preprocessing
Annotation profiles were not found in `reference.varm["group"]`. Constructing reference profiles with `tacco.preprocessing.construct_reference_profiles` and default arguments...
Finished preprocessing in 0.42 seconds.
Starting annotation of data with shape (1500, 19737) and a reference of shape (1500, 19737) using the following wrapped method:
+- platform normalization: platform_iterations=0, gene_keys=group, normalize_to=adata
+- multi center: multi_center=10 multi_center_amplitudes=True
+- bisection boost: bisections=4, bisection_divisor=3
+- core: method=OT annotation_prior=None
mean,std( rescaling(gene) ) 0.37671331182229645 0.19547104710406613
bisection run on 1
bisection run on 0.6666666666666667
bisection run on 0.4444444444444444
bisection run on 0.2962962962962963
bisection run on 0.19753086419753085
bisection run on 0.09876543209876543
Finished annotation in 7.44 seconds.
[8]:
AnnData object with n_obs × n_vars = 1500 × 25000
obs: 'group', 'libsize', 'has_program', 'program_usage', 'is_doublet', 'group2'
var: 'BaseGeneMean', 'is_outlier', 'outlier_ratio', 'gene_mean', 'prog_gene', 'prog_genemean', 'group1_DEratio', 'group1_genemean', 'group2_DEratio', 'group2_genemean', 'group3_DEratio', 'group3_genemean', 'group4_DEratio', 'group4_genemean', 'group5_DEratio', 'group5_genemean', 'group6_DEratio', 'group6_genemean', 'group7_DEratio', 'group7_genemean', 'group8_DEratio', 'group8_genemean', 'group9_DEratio', 'group9_genemean', 'group10_DEratio', 'group10_genemean', 'group11_DEratio', 'group11_genemean', 'group12_DEratio', 'group12_genemean', 'group13_DEratio', 'group13_genemean'
uns: 'TACCO_mc10'
obsm: 'TACCO_mc10', 'TACCO'
varm: 'TACCO_mc10'
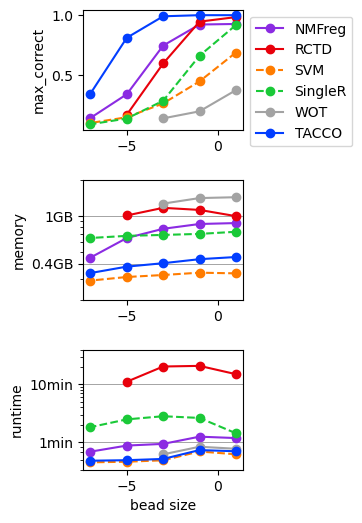
Benchmark annotation methods on the simulated dropout data¶
Define parameters for the annotation methods to use
[9]:
methods = {
'NMFreg':{'method': 'NMFreg',},
'RCTD':{'method': 'RCTD', 'conda_env': f'{env_path}/RCTD_env',},
'SVM':{'method':'svm',},
'SingleR':{'method':'SingleR', 'conda_env': f'{env_path}/SingleR_env',},
'WOT':{'method': 'WOT',},
'TACCO': {'method': 'OT', 'multi_center': 10,},
}
[10]:
results = {}
for dropout_midpoint in dropout_midpoints:
for method,params in methods.items():
print(f'running method {method} for dropout midpoint {dropout_midpoint} ...', end='')
try:
results[(dropout_midpoint,method)] = tc.benchmarking.benchmark_annotate(sdatas[dropout_midpoint],reference,annotation_key='group',**params, assume_valid_counts=True);
except:
pass # catch failing methods
print(f'done')
running method NMFreg for dropout midpoint 1.0 ...done
running method RCTD for dropout midpoint 1.0 ...done
running method SVM for dropout midpoint 1.0 ...done
running method SingleR for dropout midpoint 1.0 ...done
running method WOT for dropout midpoint 1.0 ...done
running method TACCO for dropout midpoint 1.0 ...done
running method NMFreg for dropout midpoint -1.0 ...done
running method RCTD for dropout midpoint -1.0 ...done
running method SVM for dropout midpoint -1.0 ...done
running method SingleR for dropout midpoint -1.0 ...done
running method WOT for dropout midpoint -1.0 ...done
running method TACCO for dropout midpoint -1.0 ...done
running method NMFreg for dropout midpoint -3.0 ...done
running method RCTD for dropout midpoint -3.0 ...done
running method SVM for dropout midpoint -3.0 ...done
running method SingleR for dropout midpoint -3.0 ...done
running method WOT for dropout midpoint -3.0 ...done
running method TACCO for dropout midpoint -3.0 ...done
running method NMFreg for dropout midpoint -5.0 ...done
running method RCTD for dropout midpoint -5.0 ...done
running method SVM for dropout midpoint -5.0 ...done
running method SingleR for dropout midpoint -5.0 ...done
running method WOT for dropout midpoint -5.0 ...Starting preprocessing
Finished preprocessing in 0.39 seconds.
Starting annotation of data with shape (1500, 13619) and a reference of shape (1500, 13619) using the following wrapped method:
+- core: method=WOT annotation_prior=None
Exception in WaddingtonOT. Trying with cell normalization.
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/anndata/_core/anndata.py:1828: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:134: RuntimeWarning: divide by zero encountered in true_divide
b = (q / (K.T.dot(np.multiply(a, dx)))) ** alpha2 * np.exp(-v / (lambda2 + epsilon_i))
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:138: RuntimeWarning: divide by zero encountered in log
u = u + epsilon_i * np.log(a)
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:140: RuntimeWarning: invalid value encountered in add
K = np.exp((np.array([u]).T - C + np.array([v])) / epsilon_i)
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/anndata/_core/anndata.py:1828: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:159: RuntimeWarning: invalid value encountered in double_scalars
np.linalg.norm(_a - old_a * np.exp(u / epsilon_i)) / (1 + np.linalg.norm(_a)),
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:148: RuntimeWarning: overflow encountered in exp
_a = a * np.exp(u / epsilon_i)
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:159: RuntimeWarning: overflow encountered in exp
np.linalg.norm(_a - old_a * np.exp(u / epsilon_i)) / (1 + np.linalg.norm(_a)),
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:159: RuntimeWarning: invalid value encountered in subtract
np.linalg.norm(_a - old_a * np.exp(u / epsilon_i)) / (1 + np.linalg.norm(_a)),
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:133: RuntimeWarning: overflow encountered in true_divide
a = (p / (K.dot(np.multiply(b, dy)))) ** alpha1 * np.exp(-u / (lambda1 + epsilon_i))
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:139: RuntimeWarning: divide by zero encountered in log
v = v + epsilon_i * np.log(b) # absorb
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:140: RuntimeWarning: invalid value encountered in add
K = np.exp((np.array([u]).T - C + np.array([v])) / epsilon_i)
Traceback (most recent call last):
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_wot.py", line 42, in _annotate_wot
tmap_annotated = ot_model.compute_transport_map(0, 1)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/ot_model.py", line 231, in compute_transport_map
return self.compute_single_transport_map(config)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/ot_model.py", line 304, in compute_single_transport_map
tmap, learned_growth = wot.ot.compute_transport_matrix(solver=self.solver, **config)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py", line 30, in compute_transport_matrix
tmap = solver(**params)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py", line 163, in optimal_transport_duality_gap
raise RuntimeError("Overflow encountered in duality gap computation, please report this incident")
RuntimeError: Overflow encountered in duality gap computation, please report this incident
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/workflow/simulated_dropout/temp_benchmark_sntgs8wj/benchmark_script_bxfknfzf.py", line 22, in <module>
result = tc.tl.annotate(adata, reference, **kw_args)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_annotate.py", line 802, in annotate
cell_type = _method(tdata, reference, annotation_key, annotation_prior, verbose)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_annotate.py", line 119, in _method
cell_type = annotate(adata, reference, annotation_key, **verbose_arg, **kw_args)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_wot.py", line 47, in _annotate_wot
tmap_annotated = ot_model.compute_transport_map(0, 1)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/ot_model.py", line 231, in compute_transport_map
return self.compute_single_transport_map(config)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/ot_model.py", line 304, in compute_single_transport_map
tmap, learned_growth = wot.ot.compute_transport_matrix(solver=self.solver, **config)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py", line 30, in compute_transport_matrix
tmap = solver(**params)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py", line 163, in optimal_transport_duality_gap
raise RuntimeError("Overflow encountered in duality gap computation, please report this incident")
RuntimeError: Overflow encountered in duality gap computation, please report this incident
wall_clock_time_seconds 12.60
max_memory_used_kbytes 1066564
exit_status 1
done
running method TACCO for dropout midpoint -5.0 ...done
running method NMFreg for dropout midpoint -7.0 ...done
running method RCTD for dropout midpoint -7.0 ...Starting preprocessing
Finished preprocessing in 0.33 seconds.
Starting annotation of data with shape (1483, 6771) and a reference of shape (1500, 6771) using the following wrapped method:
+- core: method=RCTD annotation_prior=None conda_env=/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/results/env_links/RCTD_env
[1] "reference.h5ad" "data.h5ad" "result" "0"
[5] "8" "full" "group" "x"
[9] "y" "7.0"
[1] "reading data"
[1] "csr"
[1] "reading reference"
[1] "csr"
[1] "running RCTD"
1 2 3 4 5 6 7 8 9 10 11 12 13
110 122 102 112 112 132 110 122 108 116 120 112 121
Attaching package: 'SparseM'
The following object is masked from 'package:base':
backsolve
Begin: process_cell_type_info
process_cell_type_info: number of cells in reference: 1499
process_cell_type_info: number of genes in reference: 6771
End: process_cell_type_info
create.RCTD: getting regression differentially expressed genes:
get_de_genes: 1 found DE genes: 31
get_de_genes: 2 found DE genes: 25
get_de_genes: 3 found DE genes: 26
get_de_genes: 4 found DE genes: 18
get_de_genes: 5 found DE genes: 25
get_de_genes: 6 found DE genes: 22
get_de_genes: 7 found DE genes: 16
get_de_genes: 8 found DE genes: 23
get_de_genes: 9 found DE genes: 29
get_de_genes: 10 found DE genes: 18
get_de_genes: 11 found DE genes: 18
get_de_genes: 12 found DE genes: 19
get_de_genes: 13 found DE genes: 16
get_de_genes: total DE genes: 260
create.RCTD: getting platform effect normalization differentially expressed genes:
get_de_genes: 1 found DE genes: 54
get_de_genes: 2 found DE genes: 42
get_de_genes: 3 found DE genes: 62
get_de_genes: 4 found DE genes: 40
get_de_genes: 5 found DE genes: 56
get_de_genes: 6 found DE genes: 44
get_de_genes: 7 found DE genes: 35
get_de_genes: 8 found DE genes: 48
get_de_genes: 9 found DE genes: 49
get_de_genes: 10 found DE genes: 48
get_de_genes: 11 found DE genes: 43
get_de_genes: 12 found DE genes: 28
get_de_genes: 13 found DE genes: 38
get_de_genes: total DE genes: 440
fitBulk: decomposing bulk
Error in choose_sigma_c(RCTD) :
choose_sigma_c determined a N_fit of 0! This is probably due to unusually low UMI counts per bead in your dataset. Try decreasing the parameter UMI_min_sigma. It currently is 7.0 but none of the beads had counts larger than that.
Calls: run.RCTD -> choose_sigma_c
Execution halted
/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_RCTD.py:239: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
adata.obs[x_coord_name] = 0
Traceback (most recent call last):
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/workflow/simulated_dropout/temp_benchmark_yz4_fh5y/benchmark_script_jq4d9rx5.py", line 22, in <module>
result = tc.tl.annotate(adata, reference, **kw_args)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_annotate.py", line 802, in annotate
cell_type = _method(tdata, reference, annotation_key, annotation_prior, verbose)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_annotate.py", line 119, in _method
cell_type = annotate(adata, reference, annotation_key, **verbose_arg, **kw_args)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_RCTD.py", line 273, in annotate_RCTD
raise Exception('RCTD did not work properly!')
Exception: RCTD did not work properly!
wall_clock_time_seconds 18.20
max_memory_used_kbytes 510112
exit_status 1
done
running method SVM for dropout midpoint -7.0 ...done
running method SingleR for dropout midpoint -7.0 ...done
running method WOT for dropout midpoint -7.0 ...Starting preprocessing
Finished preprocessing in 0.33 seconds.
Starting annotation of data with shape (1483, 6771) and a reference of shape (1500, 6771) using the following wrapped method:
+- core: method=WOT annotation_prior=None
Exception in WaddingtonOT. Trying with cell normalization.
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/anndata/_core/anndata.py:1828: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:134: RuntimeWarning: divide by zero encountered in true_divide
b = (q / (K.T.dot(np.multiply(a, dx)))) ** alpha2 * np.exp(-v / (lambda2 + epsilon_i))
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:138: RuntimeWarning: divide by zero encountered in log
u = u + epsilon_i * np.log(a)
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:140: RuntimeWarning: invalid value encountered in add
K = np.exp((np.array([u]).T - C + np.array([v])) / epsilon_i)
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/anndata/_core/anndata.py:1828: UserWarning: Observation names are not unique. To make them unique, call `.obs_names_make_unique`.
utils.warn_names_duplicates("obs")
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:159: RuntimeWarning: invalid value encountered in double_scalars
np.linalg.norm(_a - old_a * np.exp(u / epsilon_i)) / (1 + np.linalg.norm(_a)),
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:148: RuntimeWarning: overflow encountered in exp
_a = a * np.exp(u / epsilon_i)
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:159: RuntimeWarning: overflow encountered in exp
np.linalg.norm(_a - old_a * np.exp(u / epsilon_i)) / (1 + np.linalg.norm(_a)),
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:159: RuntimeWarning: invalid value encountered in subtract
np.linalg.norm(_a - old_a * np.exp(u / epsilon_i)) / (1 + np.linalg.norm(_a)),
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:133: RuntimeWarning: divide by zero encountered in true_divide
a = (p / (K.dot(np.multiply(b, dy)))) ** alpha1 * np.exp(-u / (lambda1 + epsilon_i))
/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py:133: RuntimeWarning: overflow encountered in true_divide
a = (p / (K.dot(np.multiply(b, dy)))) ** alpha1 * np.exp(-u / (lambda1 + epsilon_i))
Traceback (most recent call last):
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_wot.py", line 42, in _annotate_wot
tmap_annotated = ot_model.compute_transport_map(0, 1)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/ot_model.py", line 231, in compute_transport_map
return self.compute_single_transport_map(config)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/ot_model.py", line 304, in compute_single_transport_map
tmap, learned_growth = wot.ot.compute_transport_matrix(solver=self.solver, **config)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py", line 30, in compute_transport_matrix
tmap = solver(**params)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py", line 163, in optimal_transport_duality_gap
raise RuntimeError("Overflow encountered in duality gap computation, please report this incident")
RuntimeError: Overflow encountered in duality gap computation, please report this incident
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/workflow/simulated_dropout/temp_benchmark_53yspg0j/benchmark_script_k4mku6of.py", line 22, in <module>
result = tc.tl.annotate(adata, reference, **kw_args)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_annotate.py", line 802, in annotate
cell_type = _method(tdata, reference, annotation_key, annotation_prior, verbose)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_annotate.py", line 119, in _method
cell_type = annotate(adata, reference, annotation_key, **verbose_arg, **kw_args)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco/tacco/tools/_wot.py", line 47, in _annotate_wot
tmap_annotated = ot_model.compute_transport_map(0, 1)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/ot_model.py", line 231, in compute_transport_map
return self.compute_single_transport_map(config)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/ot_model.py", line 304, in compute_single_transport_map
tmap, learned_growth = wot.ot.compute_transport_matrix(solver=self.solver, **config)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py", line 30, in compute_transport_matrix
tmap = solver(**params)
File "/ahg/regevdata/projects/mouse_CRC/rerun/tacco_examples_devel/.snakemake/conda/1d2a210995279fd3d78066142f3cf09b/lib/python3.9/site-packages/wot/ot/optimal_transport.py", line 163, in optimal_transport_duality_gap
raise RuntimeError("Overflow encountered in duality gap computation, please report this incident")
RuntimeError: Overflow encountered in duality gap computation, please report this incident
wall_clock_time_seconds 10.58
max_memory_used_kbytes 725876
exit_status 1
done
running method TACCO for dropout midpoint -7.0 ...done
[11]:
for (dropout_midpoint,method),result in results.items():
unused_key = tc.utils.find_unused_key(sdatas[dropout_midpoint].obsm)
sdatas[dropout_midpoint].obsm[unused_key] = results[(dropout_midpoint,method)]['annotation']
max_correct = tc.ev.compute_err(sdatas[dropout_midpoint], unused_key, 'group', err_method='max_correct')[unused_key]
del sdatas[dropout_midpoint].obsm[unused_key]
results[(dropout_midpoint,method)]['max_correct'] = max_correct
[12]:
res_df = pd.DataFrame([
[dropout_midpoint,method,v['max_correct'],v['max_mem_usage_GB'],v['benchmark_time_s']]
for (dropout_midpoint,method),v in results.items()
],columns=['dropout_midpoint','method','max_correct','memory (GB)','time (s)'])
quantities = [c for c in res_df.columns if c not in ['dropout_midpoint','method'] ]
methods = res_df['method'].unique()
[13]:
fig,axs = tc.pl.subplots(1,len(quantities), axsize=np.array([4,3])*0.4, x_padding=0.7, y_padding=0.5)
colors = {m:common_code.method_color(m) for m in methods}
styles = {m:common_code.method_style(m) for m in methods}
res_df = res_df.loc[~res_df[quantities].isna().all(axis=1)]
for iy_ax, qty in enumerate(quantities):
ax = axs[iy_ax,0]
x = res_df['dropout_midpoint']
y = res_df[qty]
if qty == 'time (s)': # part 1 of adding second, minute and hour marker: plot the lines under the data
ynew = np.array([0.1,1,10,60,600,3600,36000])
ynew_minor = np.concatenate([np.arange(0.1,1,0.1),np.arange(1,10,1),np.arange(10,60,10),np.arange(60,600,60),np.arange(600,3600,600),np.arange(3600,36000,3600)]).flatten()
ynewlabels = np.array(['0.1s','1s','10s','1min','10min','1h','10h'])
ymin = y.min() * 0.5
ymax = y.max() * 2.0
ynewlabels = ynewlabels[(ynew > ymin) & (ynew < ymax)]
ynew = ynew[(ynew > ymin) & (ynew < ymax)]
ynew_minor = ynew_minor[(ynew_minor > ymin) & (ynew_minor < ymax)]
for yn in ynew:
ax.axhline(yn, color='gray', linewidth=0.5)
elif qty == 'memory (GB)':
ynew = np.array([0.1,0.4,1,4,10,40,100])
ynew_minor = np.concatenate([np.arange(0.1,1,0.1),np.arange(1,10,1),np.arange(10,100,10),np.arange(100,1000,100)]).flatten()
ynewlabels = np.array(['0.1GB','0.4GB','1GB','4GB','10GB','40GB','100GB'])
ymin = y.min() * 0.5
ymax = y.max() * 2.0
ynewlabels = ynewlabels[(ynew > ymin) & (ynew < ymax)]
ynew = ynew[(ynew > ymin) & (ynew < ymax)]
ynew_minor = ynew_minor[(ynew_minor > ymin) & (ynew_minor < ymax)]
for yn in ynew:
ax.axhline(yn, color='gray', linewidth=0.5)
for m in methods:
selector = res_df['method'] == m
if selector.sum() == 0:
continue
ax.plot(x[selector],y[selector],label=m,marker='o',color=colors[m],ls=styles[m],)
if iy_ax == axs.shape[0] - 1:
ax.set_xlabel('bead size')
if qty == 'time (s)':
ax.set_ylabel('runtime')
elif qty == 'memory (GB)':
ax.set_ylabel('memory')
else:
ax.set_ylabel(f'{qty}')
if qty in ['time (s)','memory (GB)']:
ax.set_yscale('log')
if qty in ['time (s)','memory (GB)']: # part 2 off adding second, minute and hour marker: add the second y axis after rescaling the first y axis to log scale
ax.set_yticks(ynew_minor,minor=True)
ax.set_yticks(ynew)
ax.set_yticklabels(ynewlabels)
ax.set_yticklabels([],minor=True)
if iy_ax == 0:
ax.legend(bbox_to_anchor=(1, 1), loc='upper left', ncol=1)
[ ]: